
|
|
|
Lectures
The morning sessions (Tuesday to Friday) will be devoted to the following lectures:
I. Analyzing conformational landscapes, with applications to the design of collective coordinates
II. Simulating conformational changes in proteins using meta-dynamics
III. Mining protein flexibility with robotics-inspired methods
I. Analyzing conformational landscapes, with applications to the design of collective coordinates
Instructors:
- Frédéric Cazals , INRIA Algorithms-Biology-Structure
- Charles H. Robert, Laboratory of Theoretical Biochemistry, CNRS
Topics, lecture I:
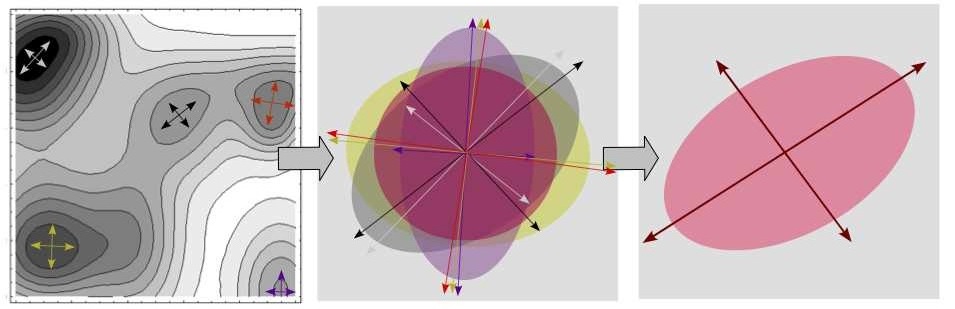
- Energy landscapes and sampling algorithms in bio-physics: overview
- Describing the topography of smooth landscapes: mathematical concepts
- Describing the topography of sampled energy landscapes: algorithms
Topics, lecture II:
- Collective coordinates, reaction coordinates, and transition paths
- Eigensystem approaches: normal modes, PCA, consensus modes
- Assessment of collective coordinates
Lecture slides :
II. Simulating conformational changes in proteins using meta-dynamics
Instructor:
Topics:
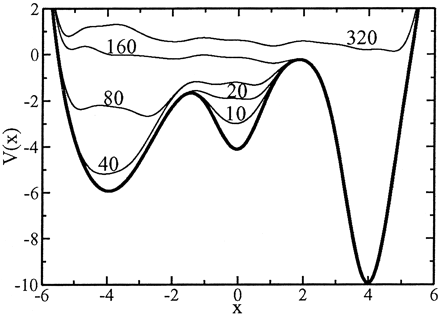
- Basic notions on free energy calculations with Metadynamics
- Best approaches to compute the free energy landscape associated with conformational changes in proteins
- How to set-up PLUMED input files
Lecture slides :
III. Mining protein flexibility with robotics-inspired methods
Instructor:
Topics:
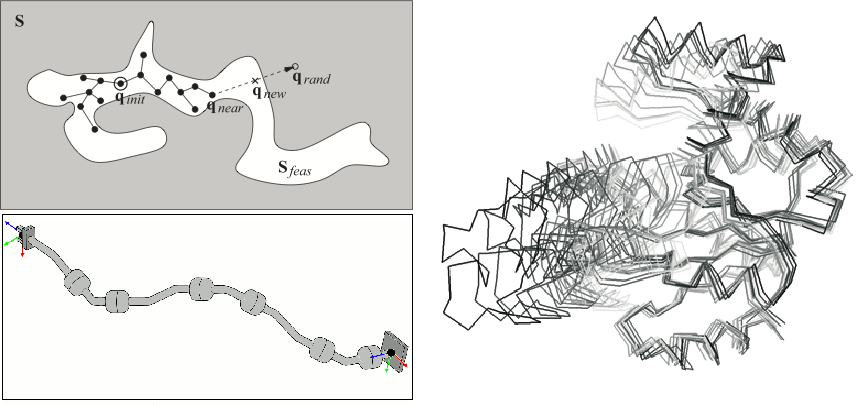
- Some basic notions on robot modeling and motion planning algorithms
- Robotics-inspired methods to explore the conformational space of proteins
- Coupling robotics algorithms and normal model analysis
Lecture slides :
IV. Determination of macromolecular structure and dynamics from experimental data:
NMR structure determination as an example
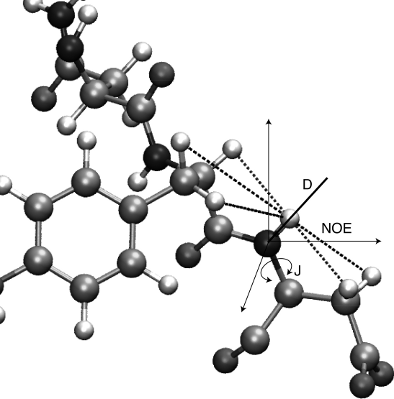
Instructor:
Topics:
- NMR data characterizing structure and dynamics of macromolecules
- Inferential structure determination
- Structure, dynamics, and interactions from NMR studies
Lecture slides :

